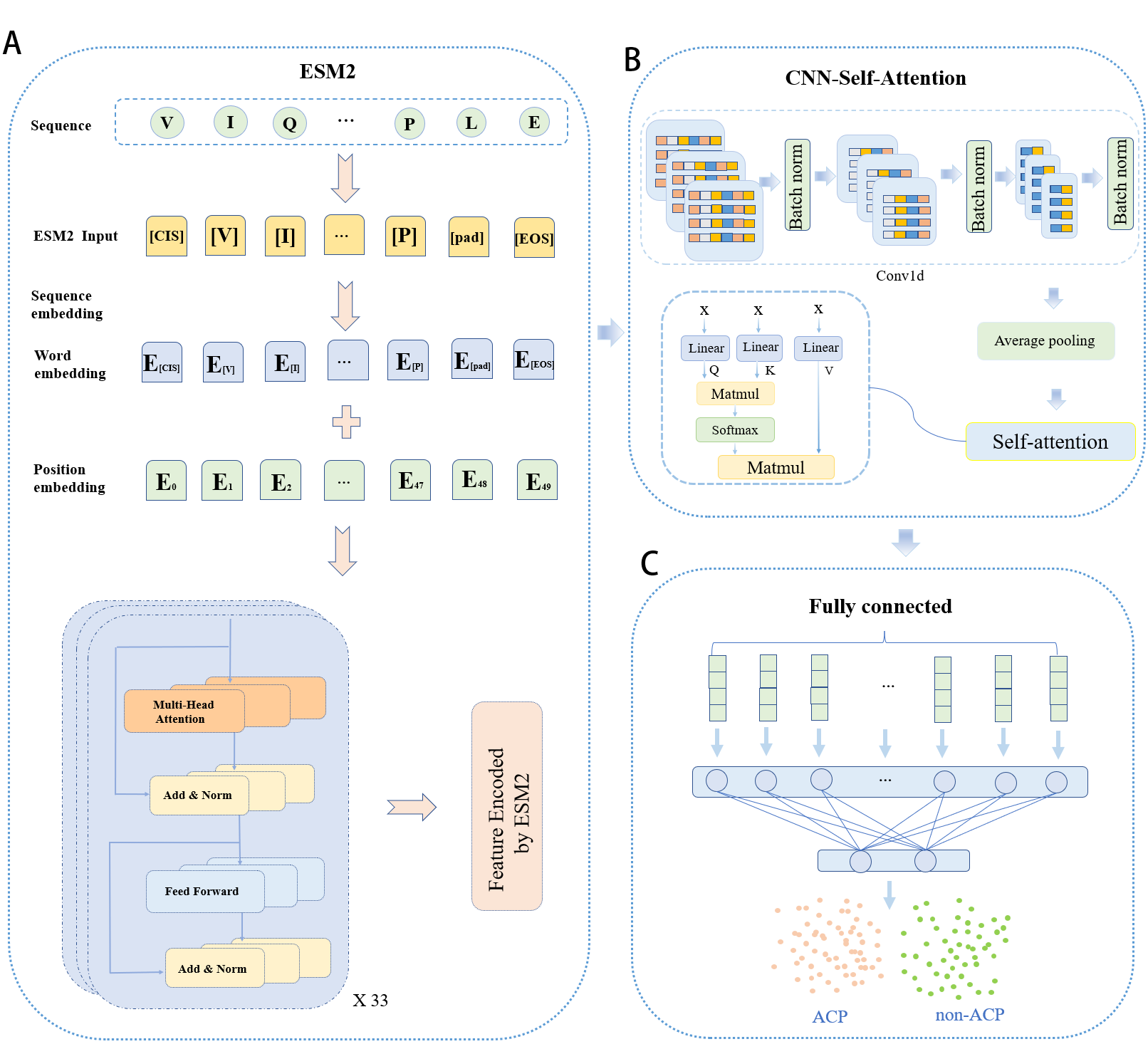
Anticancer peptides (ACPs) are short peptides with anti-cancer properties. In recent years, it has been receiving increasing attention in cancer therapy because of its small toxic, side effects and its ability to kill cancer cells accurately. However, the identification of anticancer peptides through experimental methods still requires a large investment of human resources and materials, as well as expensive and time-consuming experimental research. In addition, traditional machine learning-based ACPs prediction methods mainly rely on hand-crafted feature engineering, which usually has low prediction performance. In this study, we propose ACP-ESM2, a deep learning model framework based on Evolutionary Scale Modeling 2 (ESM2) pre-trained models. First, we treat each amino acid in the preprocessed ACPs and non-ACPs sequences as a word, which is fed into the ESM2 pre-trained model for feature extraction. In addition, the features obtained from the pre-training model ESM2 are fed into a composite model consisting of a one-dimensional convolutional neural network (CNN) and an attentional mechanism to better distinguish the features. Finally, average pooling and final classification of ACPs using fully connected layers are then performed. The experimental results show that our ACP-ESM2 predictor achieves better performance compared to existing predictors. This further demonstrates that it is effective and meaningful to capture effective feature information of peptide sequences using the ESM2 method and combine other deep learning models for ACP prediction.