Introduction
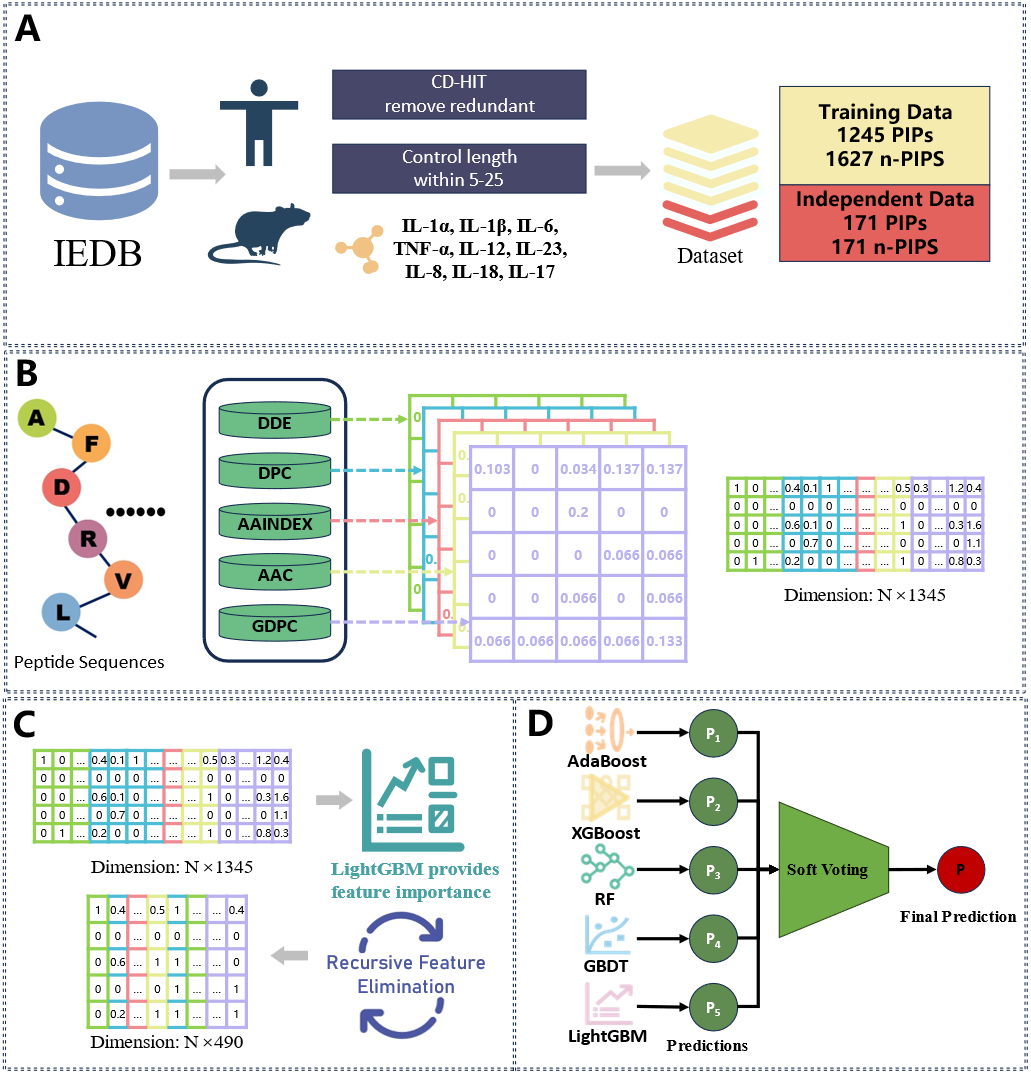
A Voting-based Ensemble Learning Framework for Predicting Proinflammatory Peptides
This framework utilizes various feature encoding techniques, such as DDE, DPC, AAINDEX, AAC, and GDPC, to represent peptide sequences. The encoded features are then processed through ensemble learning methods, including AdaBoost, XGBoost, RandomForest, GBDT, and LightGBM, to predict proinflammatory peptides.
The dataset used in this framework comprises training data of 1245 PIPs and 1627 non-PIPs, as well as independent data of 171 PIPs and 171 non-PIPs. The framework aims to enhance the prediction accuracy and provide a robust tool for bioinformatics research.