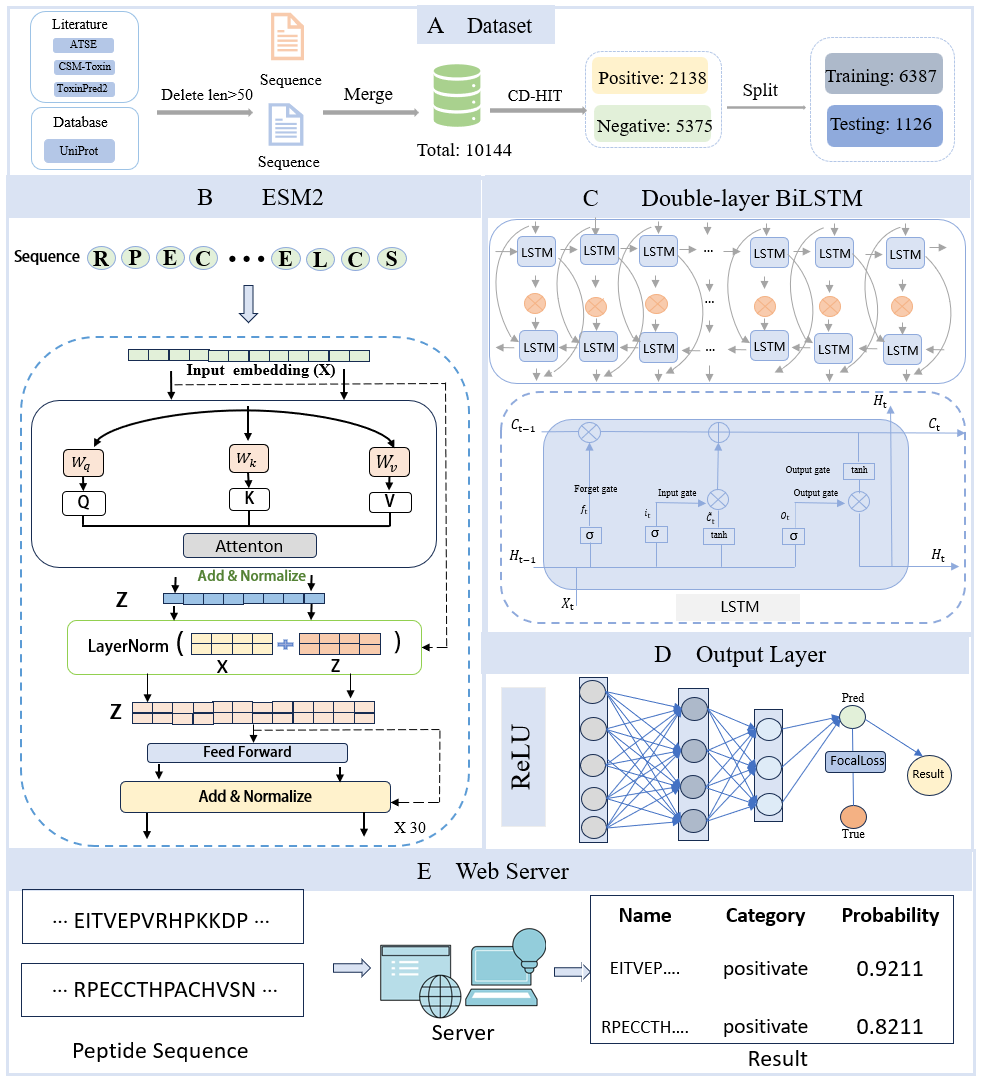
Peptide toxicity prediction holds great significance in drug development and biotechnology, as accurately identifying toxic peptide sequences aids in designing safer peptide-based drugs. This study proposes a deep learning-based model for peptide toxicity prediction, integrating Evolutionary Scale Modeling (ESM2), Bidirectional Long Short-Term Memory (BiLSTM), and Deep Neural Network (DNN). The ESM2 model captures evolutionary information from peptide sequences, while BiLSTM extracts contextual dependencies, and DNN further classifies the extracted features. To address the class imbalance in the dataset, we employed Focal Loss as the loss function to improve the identification of minority class samples. Experimental results demonstrate that the model performs exceptionally well across multiple evaluation metrics, particularly in handling imbalanced data, achieving significant improvements over traditional methods. The findings indicate that this model offers an effective solution for peptide toxicity prediction.