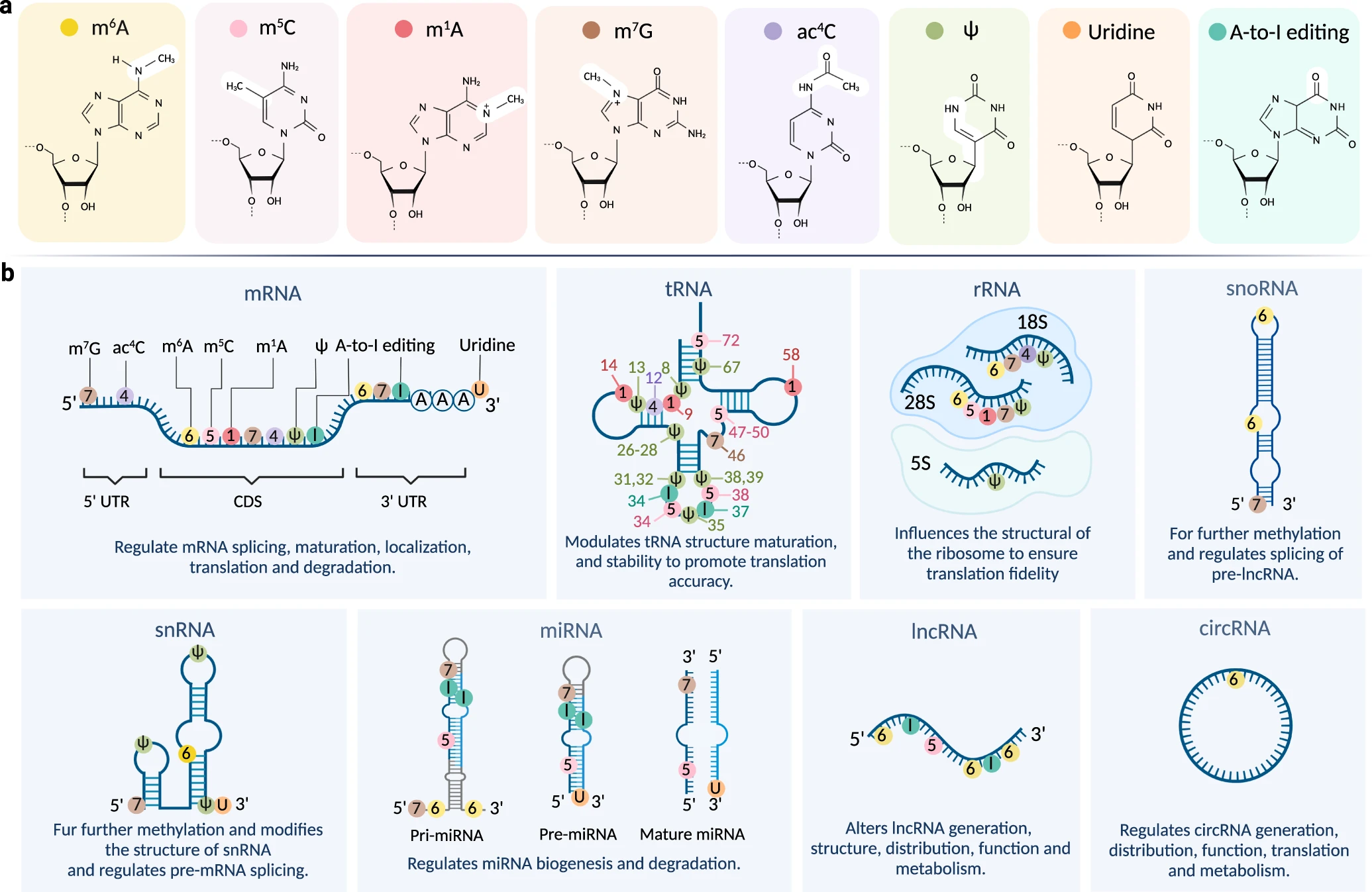
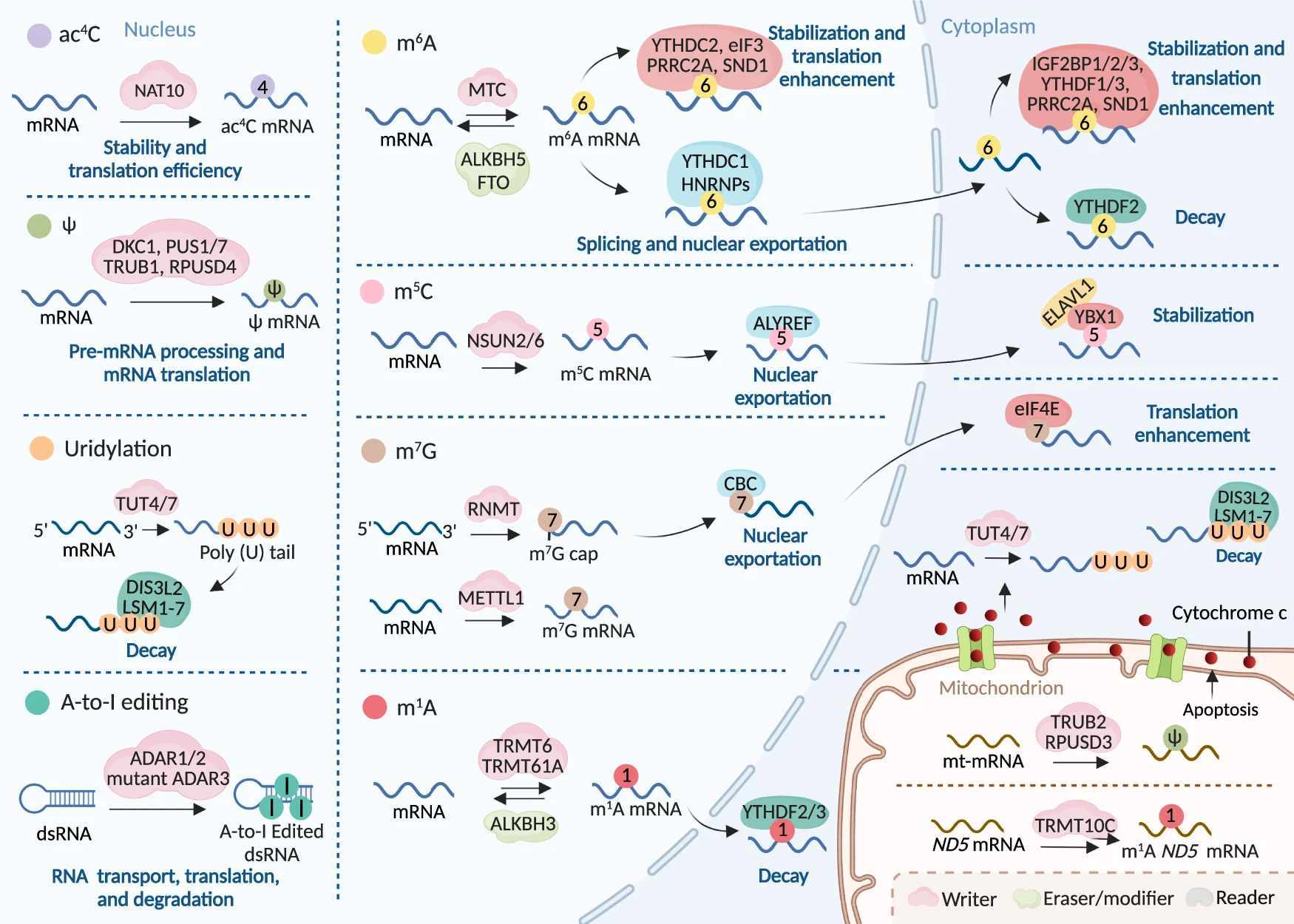
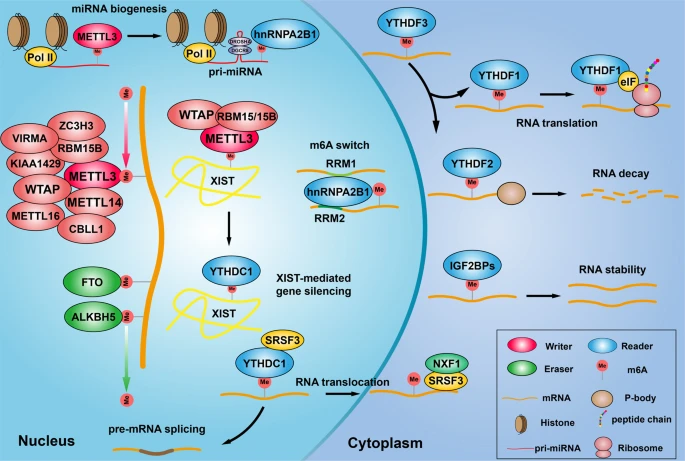
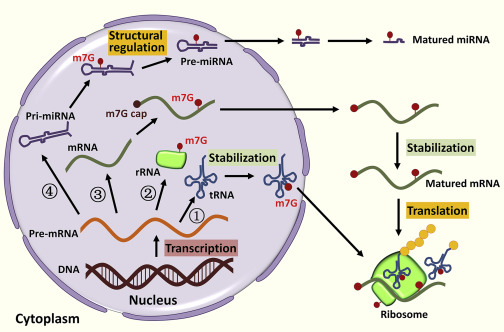
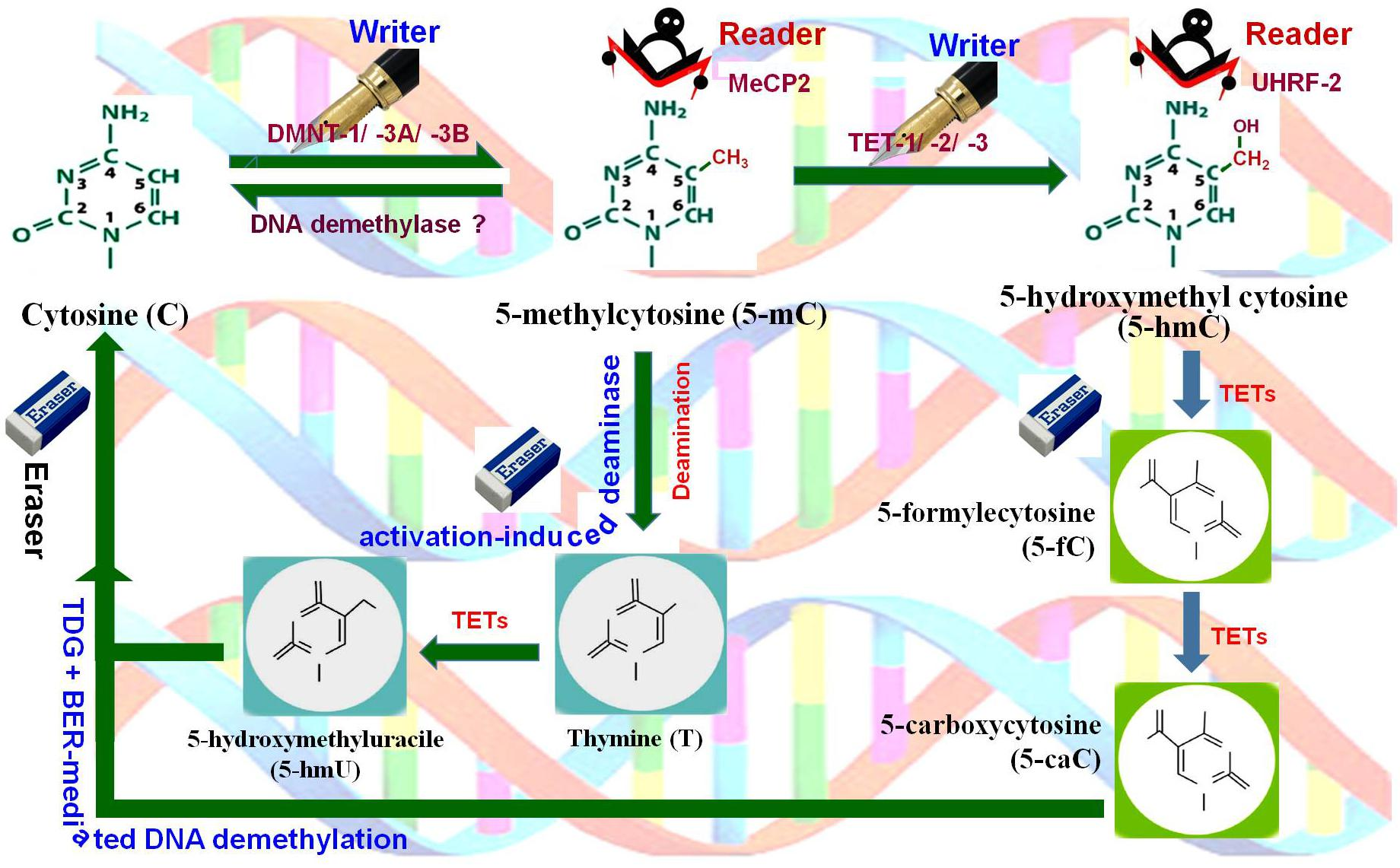
m5C
(5-Methylcytosine)
Site Function:
Gene Regulation, RNA Stability, Translation, Epigenetic Marking
5-methylcytosine (m5C) is a well-known epigenetic mark in DNA, but it also occurs in RNA, where it has distinct functions. In RNA, m5C can modulate gene expression by affecting RNA stability, translation efficiency, and the interaction with RNA-binding proteins. It is implicated in the regulation of various cellular processes, including development and disease. The presence of m5C in RNA adds another layer of complexity to the epigenetic regulation of gene expression.
Reference: Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., & Skalsky, R. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 485(7397), 201-206.