Welcome To BioAi−Lab
-
-
Hainan University, Haikou, Hainan Province, China
Welcome To BioAi−Lab
Hainan University, Haikou, Hainan Province, China
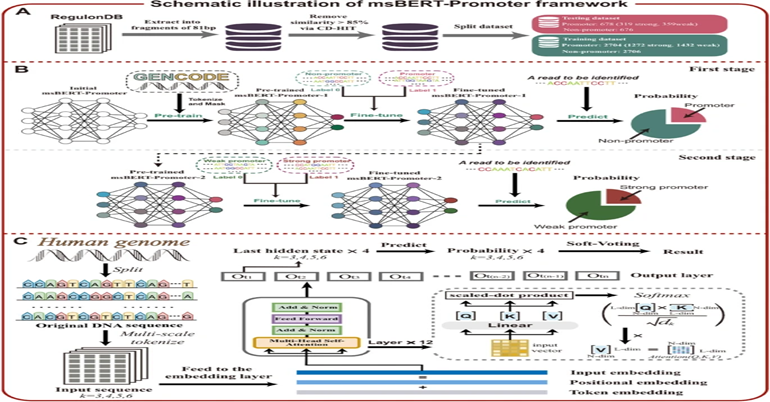
30
May
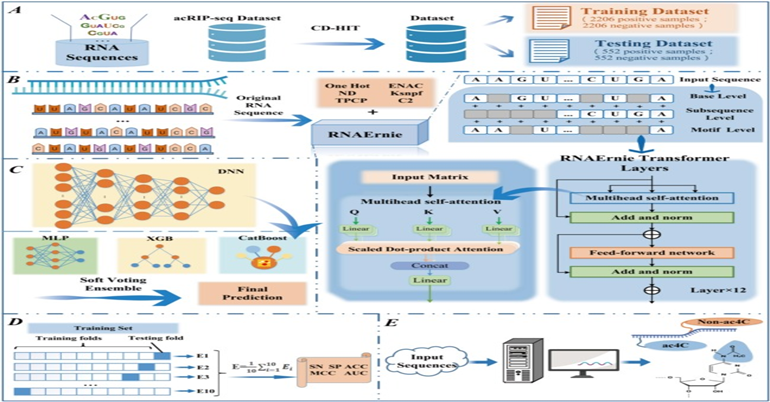
December
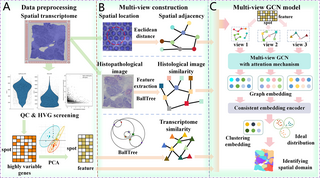
05
September
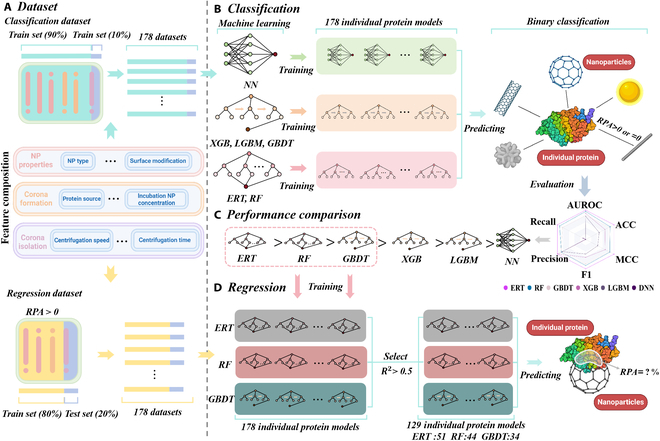
25
Sep
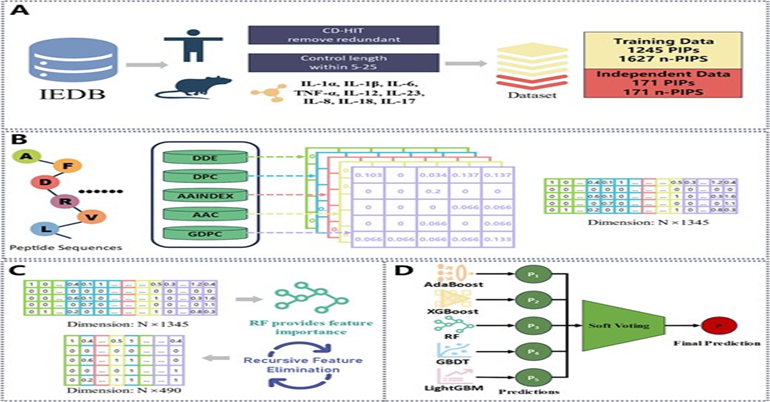
15
October
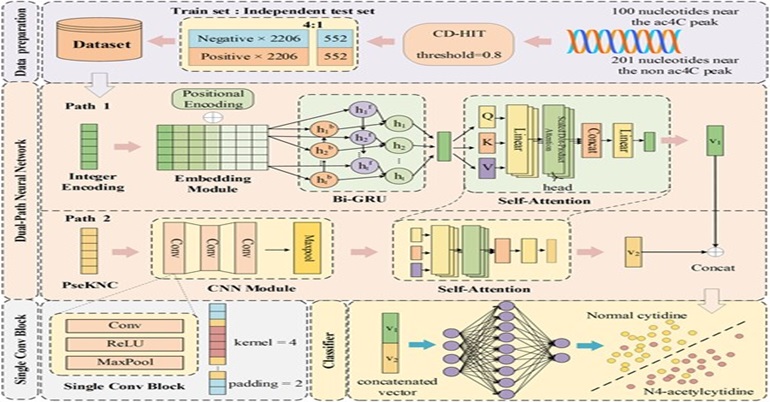
17
October

31
July
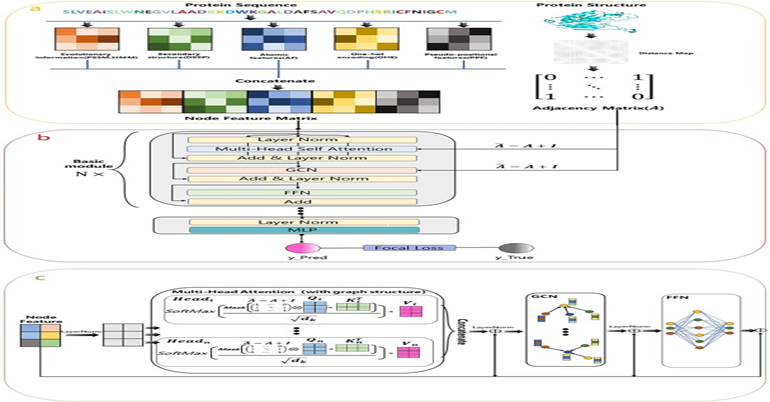
February
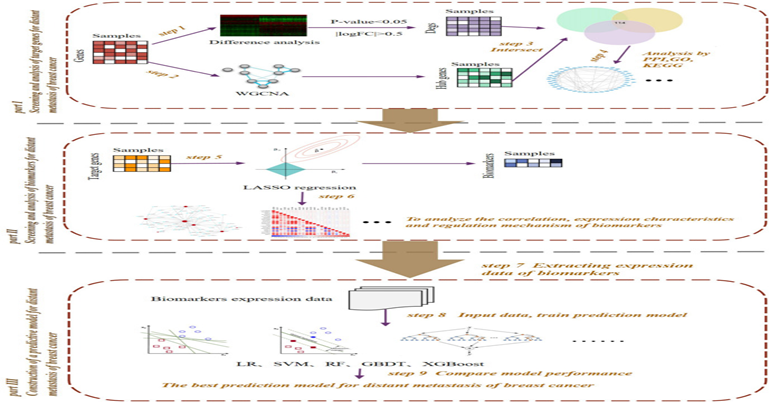
February
03
October
24
January
October
March
11
Jun

05
Jan
5-8
Dec

5-8
Dec
13
Jun
18
May
20
Apr
17
Jan
26
Nov

17
Jul

09
Mar
19
Feb
30
Jan
05
Jan
18
Dec
16
Oct